
How biomedical ontologies are unlocking the full potential of biomedical data
Our latest blog explains how SciBite's Ontologies team takes public biomedical ontologies and tailors them so that they can be used for named entity recognition (NER).

In a world where we have access to an ever-expanding sea of information – be it music, science, photos or news – curation is increasingly key. This is well-known to the major tech companies, Google, for example, employs an army of 10,000 curators (‘raters’) to assess the quality of its output.
It is especially true in the life-sciences, where the process of sifting, sorting and normalizing biomedical data (a process known as biocuration), is critical for making that data accessible and findable.
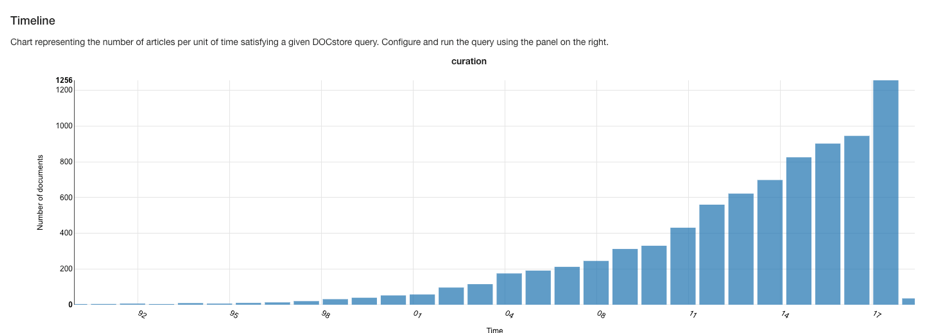
Cumulative increase in mentions of ‘curation’ in Medline since 1980. Image from DOCStore v1.3.7
Biocuration generally involves applying standard semantics – terms from biomedical ontologies and vocabularies – to data. This process may also be called ‘data annotation’. It is typically a manual process, for which professional biocurators are required to have domain expertise as well as the ability to navigate often large and complex ontologies. It is expert, labour-intensive work and as such often not scalable for large amounts of data.
This is where SciBite’s tools come in. Our Ontologies team takes public biomedical ontologies and tailors them so that they can be used for named entity recognition (NER). We do this tailoring in two key ways. First, we add more synonyms to increase the search breadth, and to allow for normalisation across entities.
For example: PDE5A, Phosphodiesterase V, Phosphodiesterase 5a, and PDEVa are all synonyms for the gene “Phosphodiesterase 5” and they all resolve to the same entity via its ID. Second, we handle ambiguity so that the matches are made only within the correct contexts, so ‘EGFR’ will match to either ‘Epidermal growth factor receptor’ or ‘e-glomerular filtration rate’ depending on the surrounding text.
This is itself a manual biocuration process, but one which allows us to scale across large amounts of information very quickly. Valuable manual effort can be reused efficiently and at scale. And while at SciBite we take pride in our manual expertise, we also super-charge work that with technologies such as machine learning and rule-based systems to make the manual work faster and more efficient.
Obviously NER can only take you so far, but once you have your entities reliably recognised and typed, you can leave your biocurators to focus on the parts that really benefit from human intervention, like extrapolating inferences from multiple sources or extracting subtle meaning from text. Alternatively, the annotated text can be used as the input to your machine learning/AI systems, helping improve performance by reducing noise and variation .
At SciBite we often build custom TERMite vocabularies for our customers, with their own data or in some specialist area, or to augment SciBite’s own vocabularies. Increasingly, we get asked to provide other specialist ontology related services such as creating new vocabularies or more formal ontologies where no public source exists, or to create custom semantic patterns to detect certain elements from text.
This is why we have decided in 2019 to launch Expert Ontology Services, which will allow our specialist team, with their many years of experience of working with ontologies, to help tackle your organisations data challenges and get you on the road to clean data.
Get in touch with the team to find out how we can work with you.
Related articles
-
Are ontologies still relevant in the age of LLMs?


Technological advancements exhibit varying degrees of longevity. Some are tried and trusted, enduring longer than others, while other technologies succumb to fleeting hype without attaining substantive fruition. One constant, in this dynamic landscape is the data.
Read -
What is Retrieval Augmented Generation and why is the data you feed it so important?


Within the life sciences, evidence-based decision-making is imperative; wrong decisions can have dire consequences. As such, it is vital that systems that support the generation and validation of hypotheses provide direct links, or provenance, to the data that was used to generate them. But how can one implement such a workflow?
Read
How could the SciBite semantic platform help you?
Get in touch with us to find out how we can transform your data
© Copyright © 2024 Elsevier Ltd., its licensors, and contributors. All rights are reserved, including those for text and data mining, AI training, and similar technologies.

