
Ontology mapping: You say tomato I say Solanum Lycopersicum…
In this post, we explore this important but often overlooked topic and discuss its relevance to the work of SciBite and other groups, such as FAIR and the Pistoia Alliance within the broader scientific community.

The problem of good ontologies
We all know ontologies are great and are the cornerstone of data sharing using FAIR principles. Ontologies describe a particular scientific domain in a way that both humans and, critically, computers can understand. At SciBite, we combine world-class ontologies with text analytics technology to transform unstructured text into rich data for analytics, mining, and deep learning activities. In an ideal world, we’d have a nice ordered set of ontologies, each covering a single, defined space. Of course, this is not an ideal world! In this space, one major challenge we face is the use of multiple ontologies covering the same type of concept.
For instance, the UBERON and FMA ontologies both cover anatomy but from different philosophical perspectives. More common are ontologies that cover multiple entity types and, thus, overlap with multiple ontologies such as NCIT, EFO and MEDDRA. Duplication across ontologies also occurs when companies have developed their own internal systems, which may cover common entities with a different perspective to those available in the public domain. Whatever the source, the issues are the same – data scientists cannot “join” data that is annotated by two separate ontologies. In order to connect the knowledge, we need to be able to bridge between, say “Asthma” in one ontology to its equivalent in another.
Ontology mapping has been a long-standing research problem in both the academic and commercial sectors, and there are many publications and software tools available. There have been many different approaches to the problem, covering a variety of algorithms and ideas.
Recently, the area has had somewhat of a boost, thanks to the Pistoia Alliance’s Ontologies Mapping project. This has brought together a good number of pharmaceutical companies, academia, and SMEs (including SciBite) under a pre-competitive framework in order to develop data, tools, and best practices around this important topic. We won’t cover the activity of this valuable project here, but we strongly suggest anyone with an interest in this topic takes a look over at their website as the team is very active and developing the next phase of their work. In this discussion, we’ll focus more on what tools SciBite has in this space.
Ontology mapping vs. Named Entity Recognition
We are often asked how ontology alignment is different from Named Entity Recognition (NER), the latter of which concerns itself with finding mentions of ontology concepts within the text – so are these two the same thing? Specifically, could we take one ontology (“source”) and load it into a NER Engine (such as SciBite’s TERMite tool) and search this against another (“target”) ontology to find similar concepts? To illustrate this, let’s take a look at a common example (concerning two ontologies chosen in the Pistoia project), the Human (HPO) and Mouse (MGI) phenotype ontologies.

Figure 1: Similar concepts found in the two major mammalian phenotype ontologies. Examples shown are part of the Pistoia Alliance Ontology Mapping Project test set.
As you can see, the first example, “increased body weight,” is represented identically in both ontologies. Thus, if we used HPO as a “source” ontology and scanned MGI as a “target”, we’d expect NER to find the same concept and produce the ontology cross-reference we need. The second example, “Obesity” vs. “Obese,” represents a non-identical match. However, a good NER system should still relate these two terms as equivalent and ignore the slight grammatical change. Things get a bit more complex in the third example, though one can make a solid argument these phrases are scientifically equivalent and perhaps should still be found by NER even though they use some different words.
When we get to example four, we’re now well into the world of subjectivity. Are these two concepts equivalent? It depends on your perspective, and, most importantly, your use case. It’s here that NER is definitely not going to help, this goes beyond its remit and requires us to use a tool that looks at the scientific meaning of these concepts to determine whether they are indeed related to each other.
One might argue that this is a problem with NER, and that we should enhance our NER engines to allow matching on the fourth example. But this would go against the real aim of NER, which is primarily to unambiguously identify mentions of specific topics in the text. While we want our NER to be flexible and match obvious permutations of synonym phrases, we don’t want it to stray too much from its original brief, for instance, deciding to match “Chronic Obstructive Pulmonary Disease” to the concept of “Asthma” just because both are inflammatory lung diseases. Thus, NER and ontology alignment can be considered to related, but quite distinct challenges. Some of the key differences are summarised in Table 1.
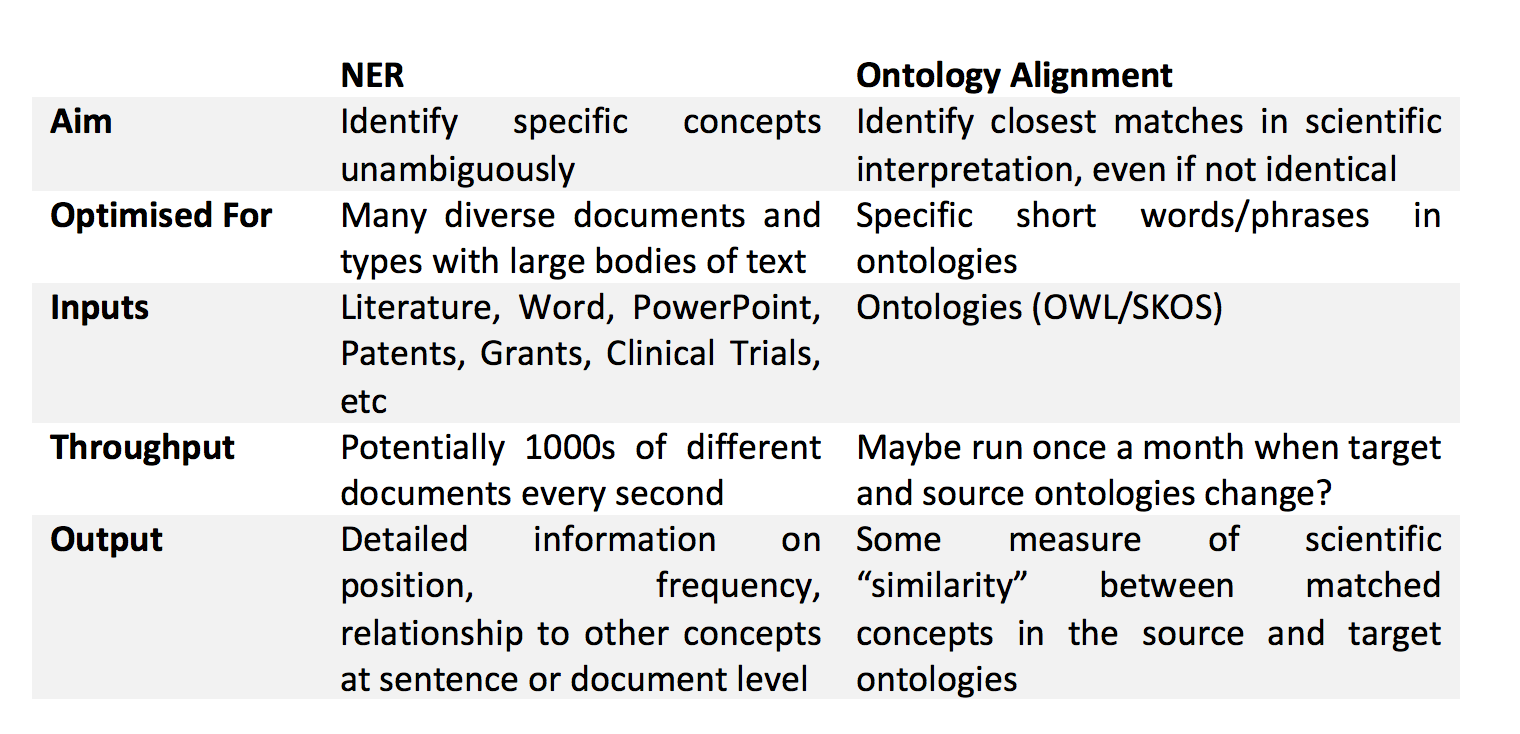
Table 1: Comparison of Named Entity Recognition and Ontology Alignment
SciBite’s Tools To The Rescue
At SciBite we’ve been working with many customers in commercial life science business to address some of the challenges above. Specifically, understanding that while ontology alignment and NER are closely related, they are distinct problems that require distinct solutions. Customers of our semantic platform have a comprehensive set of modules available to them to address any need across this spectrum:

Table 2: SciBite Tools For Ontology-based Analysis
SciBite Labs: Xlign for large-scale ontology cross-referencing
At SciBite, we’ve built an add-on module for our platform to specifically address the needs of an ontology alignment task. A product of our “SciBite Labs” initiative, where we provide customers with exclusive access to products we’re researching, Xlign (pronounced “Cross-Line”) provides a great solution to this challenge. Indeed, the tool reaches over 98% accuracy on the previously published Pistoia Ontologies Mapping test sets. Available as an API or user-facing tool, Xlign allows users to generate cross-references between any ontologies, including public and internal (proprietary) resources.
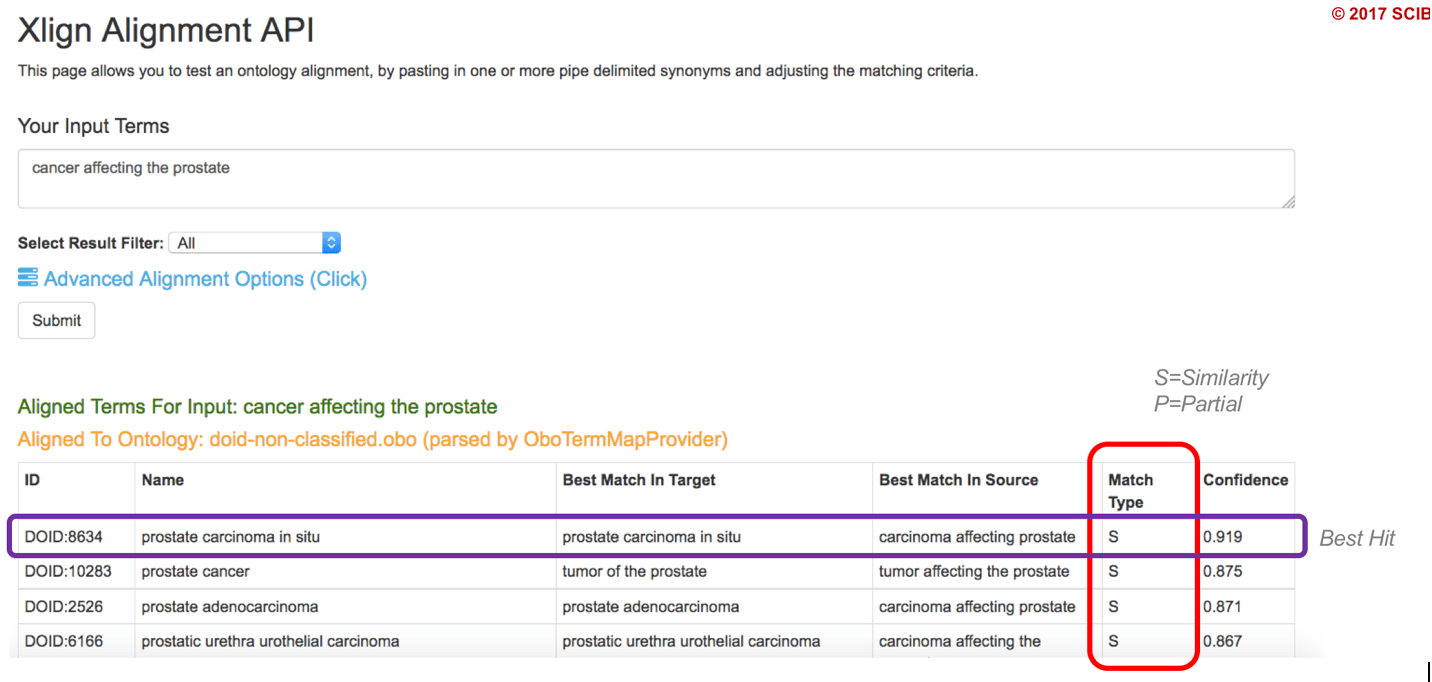
Figure 2: A proposed cross-reference by Xlign of an input term to the Human Disease Ontology.
How do we achieve such a good alignment? The basis of Xlign is no different from anything else we do at SciBite – our focus on life sciences. Over the years, we’ve built a huge amount of linguistic intelligence into our tools, specifically engineered for biology and medicine.
The core of Xlign is our award-winning TERMite software, which has been specifically tuned to relax the standard matching rules of NER and instead employs additional algorithms that scan for similarity at a scientific level. This modification allows us to compare millions of potential matches across large ontologies at high speed. By creating a matrix of all possible cross-references, then ranking these by scientific similarity, Xlign is able to identify any concepts which appear related and provide a quantifiable confidence score to allow rapid triage of the results. With options to control such as partial matching of sub-phrases, fuzzy matching for spelling variations and feature weighting, Xlign is a powerful tool for tackling the challenge of mapping scientifically similar concepts across important ontologies.
Summary
This post highlights how ontology mapping and named entity recognition are two quite different problems, both grounded in a need for high-performance scientific language processing. While NER concerns itself with finding the mention of specific things within the text, ontology mapping relaxes what is considered a “match” by looking at what may be similar, but not necessarily identical scientifically.
However, this is very subjective, and the definition of a good match will be very different between use cases and even between colleagues. Fortunately, SciBite’s TERMite and Xlign systems provide a proven solution to address these challenges, even combining with our Kusp platform for data cleansing to ensure a high-quality data pipeline from the start to finish.
Learn more about SciBite’s semantic technology platform and how we can help you get more from your data.
Related articles
-
A hacker’s guide to understanding bio-ontology jargon
Perfect for those new to bio-ontologies or who work with ontologists - a whole new vocabulary deciphered!
Read -
Creating a SciBite VOCab from a public ontology


Public ontologies are essential for applying FAIR principles to data but are not built for use in named entity recognition pipelines. At SciBite, we build on the public ontologies to create VOCabs optimized for NER. In this blog, discover how we create a SciBite VOCab from a Public Ontology.
Read
How could the SciBite semantic platform help you?
Get in touch with us to find out how we can transform your data
© Copyright © 2024 Elsevier Ltd., its licensors, and contributors. All rights are reserved, including those for text and data mining, AI training, and similar technologies.

