
Creating a SciBite VOCab from a public ontology
 |
 |
Public ontologies are essential for applying FAIR principles to data but are not built for use in named entity recognition pipelines. At SciBite, we build on the public ontologies to create VOCabs optimized for NER. In this blog, discover how we create a SciBite VOCab from a Public Ontology.

Why do we create SciBite Vocabs from public ontologies?
Publicly available ontologies and vocabularies are essential for companies who want to improve the usability of their data by applying the FAIR data principles. At SciBite, we can facilitate this process by providing expertly curated versions of public ontologies called SciBite VOCabs. These VOCabs can be run over unstructured text using TERMite, our named entity recognition (NER) software, to produce clean contextualized data harmonizing with publicly available ontology concepts.
The process of creating a SciBite VOCab from a public ontology starts with importing the public ontology into CENtree, our ontology management platform. From here, the ontology can be exported as a text file consisting of the identifiers, preferred names, and synonyms of all entities in the ontology. If we were to use this file directly for NER on unstructured text within TERMite, the output would not be optimal.
Firstly, public ontologies can sometimes lack complete coverage of a domain that a company may require; individual entities, synonyms or even whole areas of a domain may be missing. Secondly, the ontologies may also contain too much ambiguity around classes, leading to a lot of noise and incorrect hits.
High-level overview of creating a SciBite VOCab
At SciBite, we can help form better coverage and reduce ambiguity when performing NER on unstructured text by curating the public ontologies when in their SciBite VOCab formats. This helps to develop them and subsequently meet the needs of a company. Below, we provide a high-level overview of how we do this.
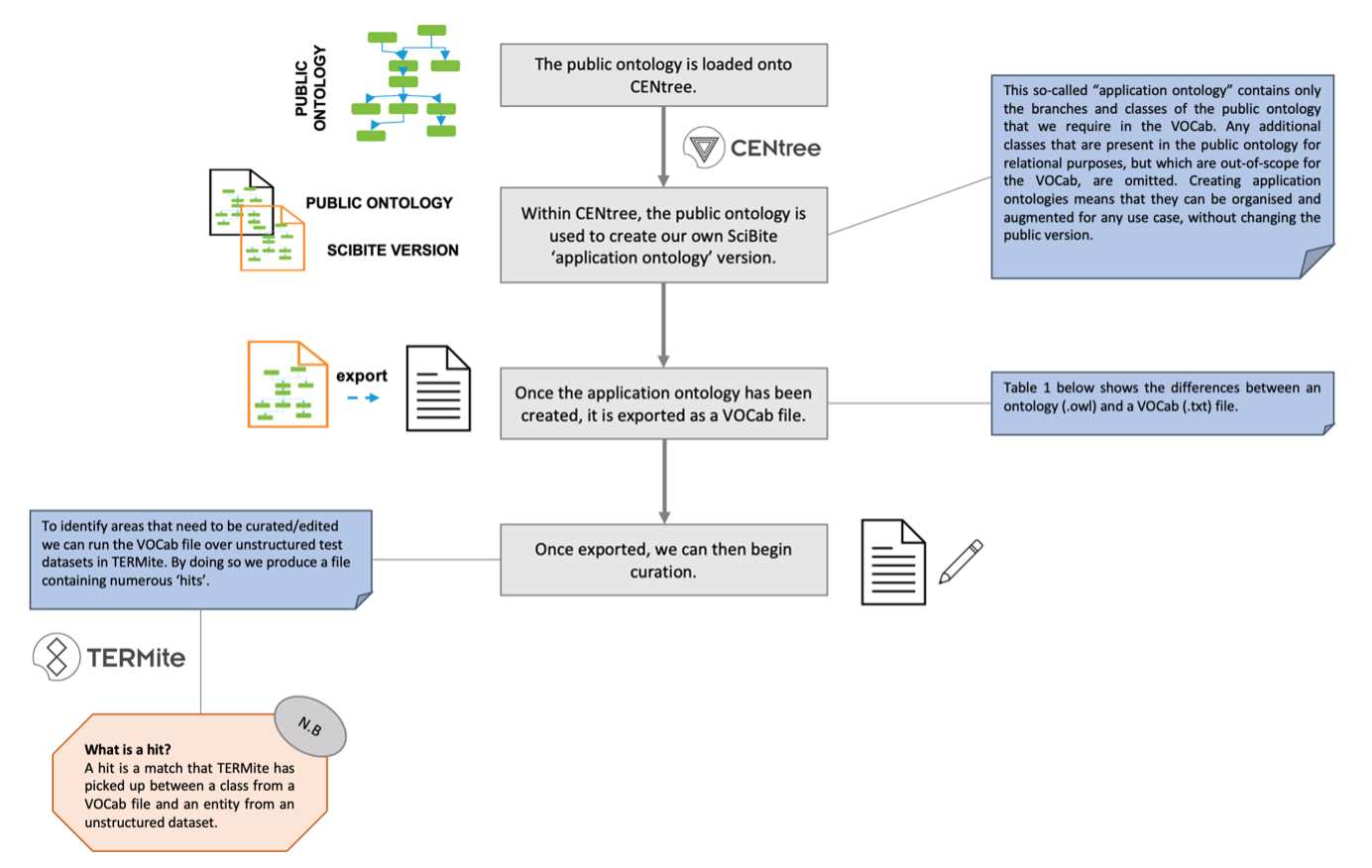
Figure 1: High-level Overview of Creating a SciBite VOCab
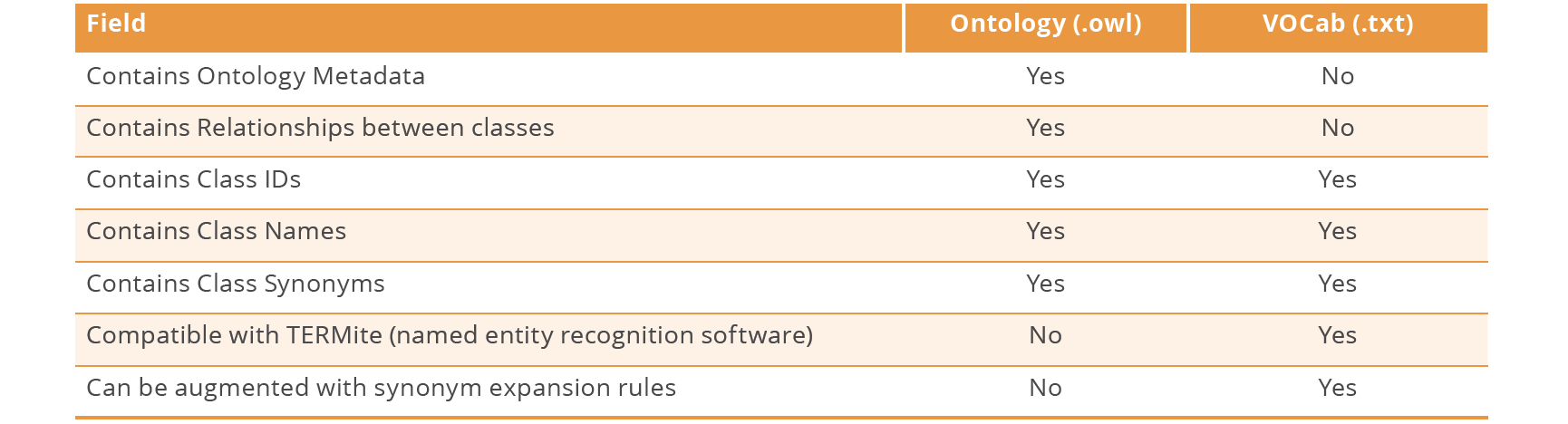
Table 1: Differences and similarities between an ontology (.owl) and a VOCab (.txt) file.
Curating the sequence ontology
Sequence Ontology (SO), to our catalog of curated vocabularies. The SO is a set of terms and relationships used to describe the features and attributes of biological sequences.
Out of the box, SO would not perform optimally in NER of unstructured text because it has ambiguously named classes, as well as missing classes and synonyms. Our expert curation team optimizes the performance of VOCabs in NER by running iterative testing of the VOCab in TERMite using scientific text followed by targeted curation.
To illustrate our new VOCab curation process, we provide a few curation cases below using SO as a typical example:
Curation Case 1 – Ambiguity
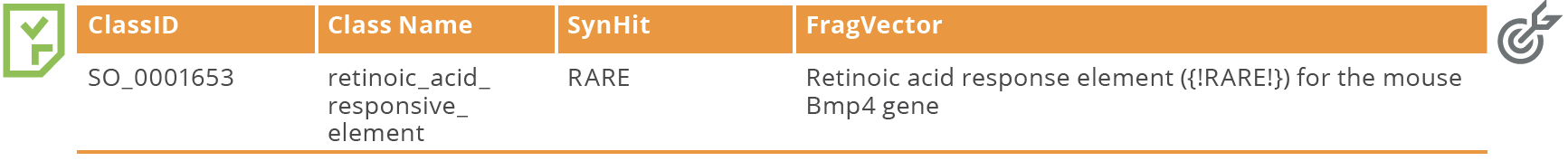
Case: After running the SO VOCab file over unstructured test dataset 1 we found multiple incorrect hits to the class SO_0001653 retinoic acid response element. As you can see below, this has happened as the term has the ambiguous synonym ‘RARE’ which is hitting to a lower case, out of context version of the word.

Fix: To reduce the noise and ambiguity that the synonym created, we curated the synonym ‘RARE’ so that it will only match in a case-sensitive manner. Therefore, entities will only hit the synonym ‘RARE’ when it is in an uppercase format, thus reducing the chances of false positive hits, such as the ‘rare diseases’ hit above.
Result: The hit is now correct for this class.
Curation Case 2 – Addition of Classes
Case: After running the SO VOCab file over unstructured test dataset 2 we found a gap in the ontology/vocabulary that we could build upon. There was a hit to the class SO_0001505 reference_genome. However, looking through the sentence (FragVector) containing the hit, we realised there was no hit to GRCh38 which is a type of reference genome. Moreover, checking the SO in full, there were no classes to represent the different types of reference genomes.
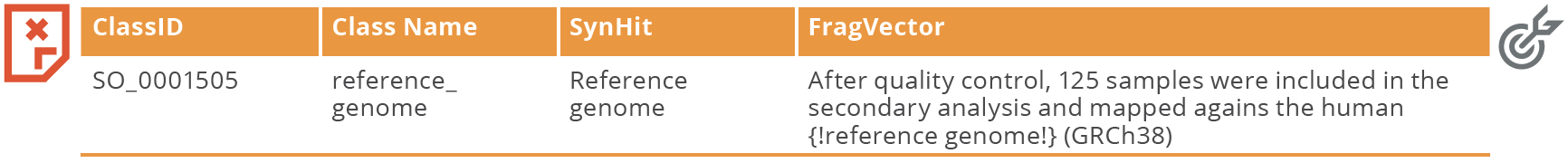
Fix: To fix this via curation, we decided to add new classes to represent the different types of reference genomes. We also added some known synonyms (syns) to the classes.
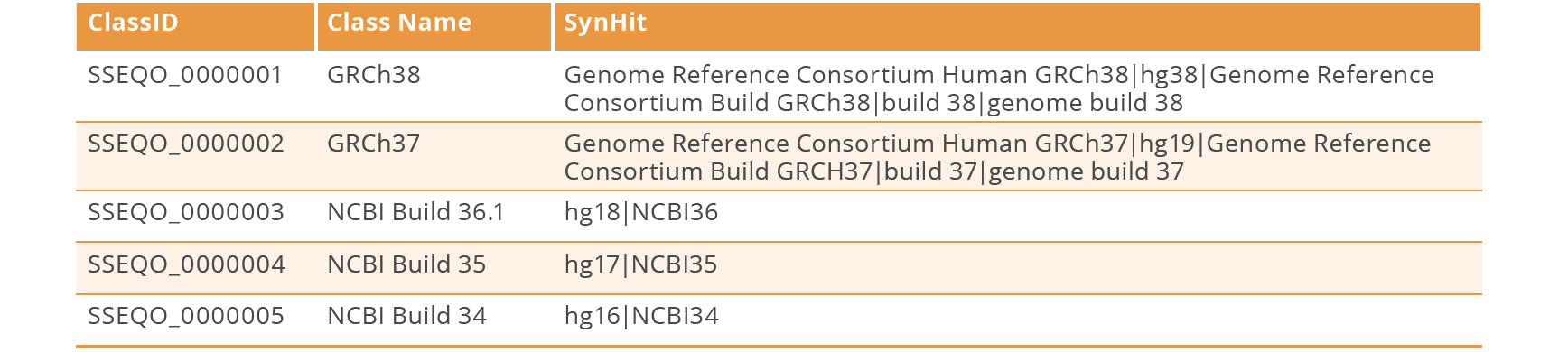
Result: The new class ‘GRCh38’ is now a hit in the text.

Curation Case 3 – Uninformative Names & Ambiguity
Case: After running the SO VOCab file over unstructured test dataset 2 we found many hits to the class SO_0000985 double. Looking into this class further, we identified that it was referring to a nucleotide polymer with two strands, thus making most of the hits incorrect due to the common use of the word ‘double’ in text (see an example below).

Fix: To fix this via curation, we firstly changed the class name to ‘double-strand’ to make it more informative. We also added synonyms such as ‘double stranded’. As the previous class name ‘double’ was too ambiguous, we also decided to remove it completely as a synonym.
Result: The hit is now correct for this class

Summary
Public ontologies are essential for applying FAIR principles to data but are not built for use in named entity recognition pipelines. At SciBite, we build on the public ontologies to create VOCabs optimised for NER. As illustrated above, each public ontology needs to undergo extensive, iterative, expert curation to provide accurate, contextual, and unambiguous matches to ontology classes in unstructured text.
SciBite is continually adding public ontologies to our catalog of curated VOCabs. If you require a specific ontology for your use case, please get in touch to discuss your requirements.
About SciBite
Our data-first, award-winning semantic analytics software is for those who want to innovate and get more from their data. Built by scientists for scientists, we believe data fuels discovery and continue to push boundaries with our cutting-edge technology applications and people-first solutions that unlock the complexities of scientific content.
Related articles
-
What’s in our 6.5.2 TERMite / VOCabs release


SciBite’s vocabularies fuel a host of use cases, from complex querying to data integration and discovery of new knowledge. In the 6.5.2 release of VOCabs, SciBite introduces the new Emtree VOCab pack, as well as a new Sequence Ontology vocab to the Genotype-Phenotype vocab pack. Several updates to existing vocabularies are also included.
Read -
Why use your ontology management platform as a central ontology server


Raw data has the inherent characteristic of being unstructured with potential quality issues such as inaccurate, incomplete, inconsistent, and duplicated. Therefore, it must be processed before it can be used for subsequent analysis and confident data-driven decisions. This is where ontologies come into play.
Read
How could the SciBite semantic platform help you?
Get in touch with us to find out how we can transform your data
© Copyright © 2024 Elsevier Ltd., its licensors, and contributors. All rights are reserved, including those for text and data mining, AI training, and similar technologies.

